传感器融合实时多目标物体对象追踪
1. 多目标追踪概述
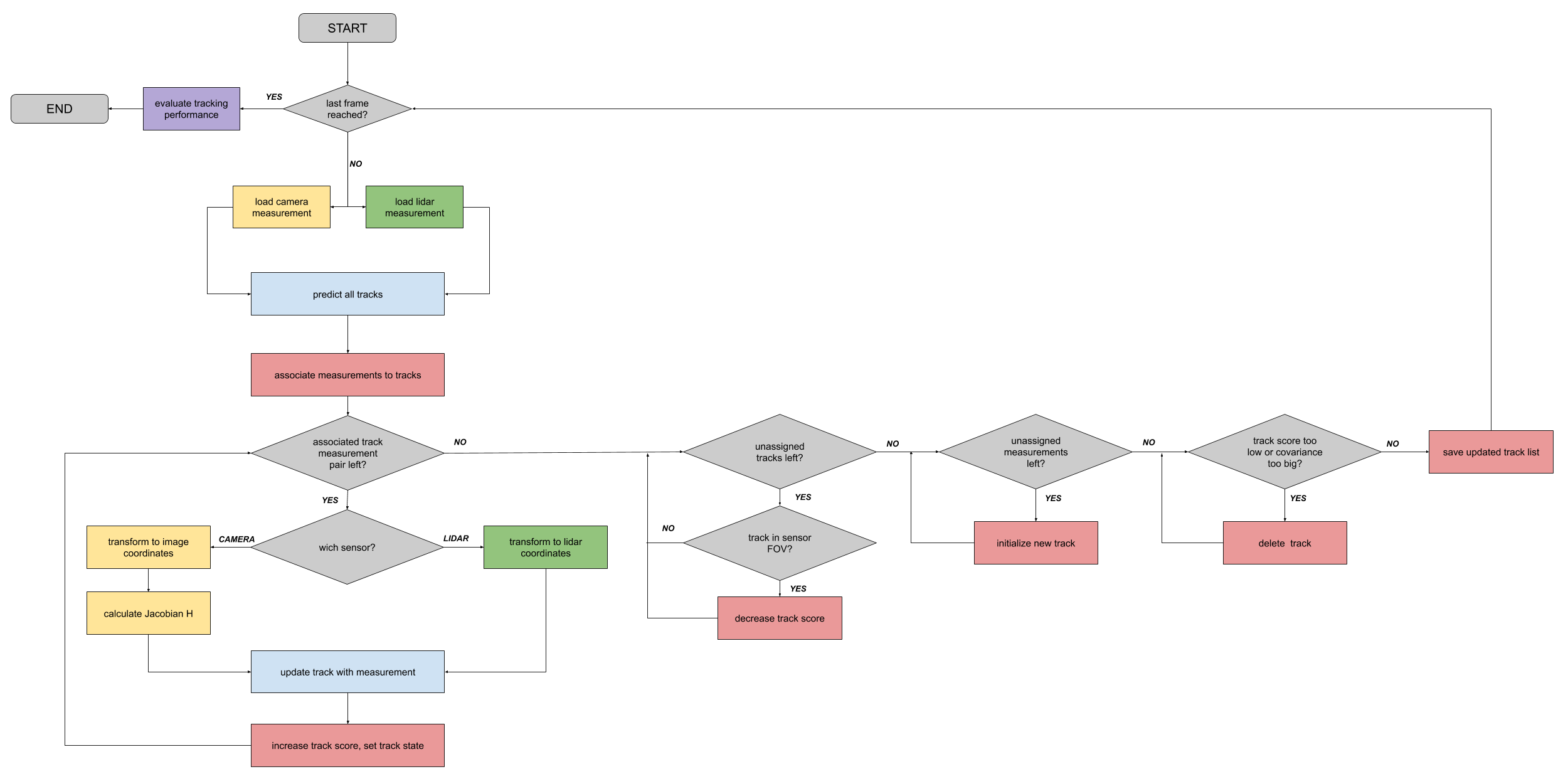
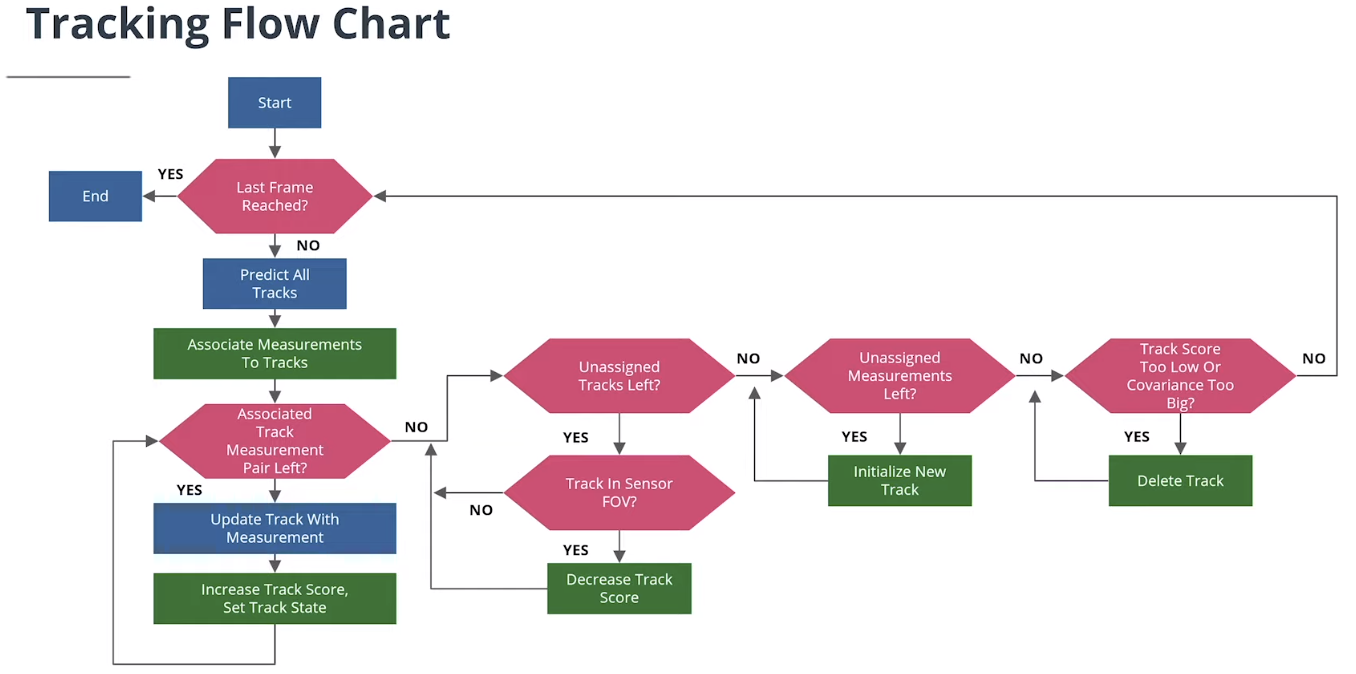
除单目标跟踪外,多目标跟踪系统还必须完成以下任务:
- 数据关联:
-
- 将测量值与轨迹关联
-
追踪管理:
-
- 初始化新跟踪对象
-
- 删除旧跟踪对象
-
- 给跟踪对象赋值置信度
2. 初始化追踪
2.1 状态初始化
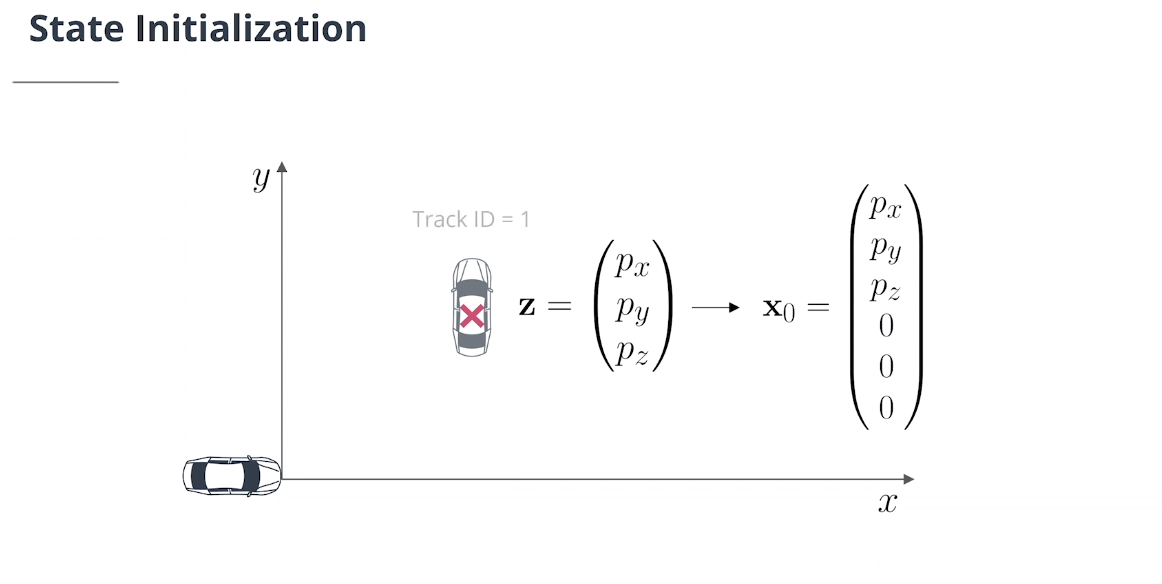
一个未分配的激光雷达测量$\mathbf z =(z_1,z_2,z_3)^ T$首先必须从传感器坐标转换为车辆坐标:
\[\begin{pmatrix} p_x\\ p_y\\p_z\\1 \end{pmatrix} = \left(\begin{array}{c|c} \begin{matrix} r_{11} & r_{12}& r_{13} \\ r_{21} & r_{22}& r_{23}\\ r_{31} & r_{32}& r_{33} \end{matrix} & \begin{matrix} t_1 \\ t_2\\ t_3 \end{matrix}\\ \overline{\begin{matrix} 0 &0&0 \end{matrix}} & \overline{\begin{matrix} 1 \end{matrix}} \end{array}\right) \cdot \begin{pmatrix} z_1\\ z_2\\z_3\\1 \end{pmatrix} = \left(\begin{array}{c|c} {\mathbf M_{\text{rot}}} & {\text t}\\ \overline{\begin{matrix} 0 &0&0 \end{matrix}} & \overline{\begin{matrix} 1 \end{matrix}} \end{array}\right) \cdot \begin{pmatrix} z_1\\ z_2\\z_3\\1 \end{pmatrix}\]然后我们可以按如下方式初始化新追踪的状态:
\[\mathbf x_0 = \begin{pmatrix} p_x\\ p_y\\ p_z\\0\\0\\0 \end{pmatrix}\]2.2 协方差初始化
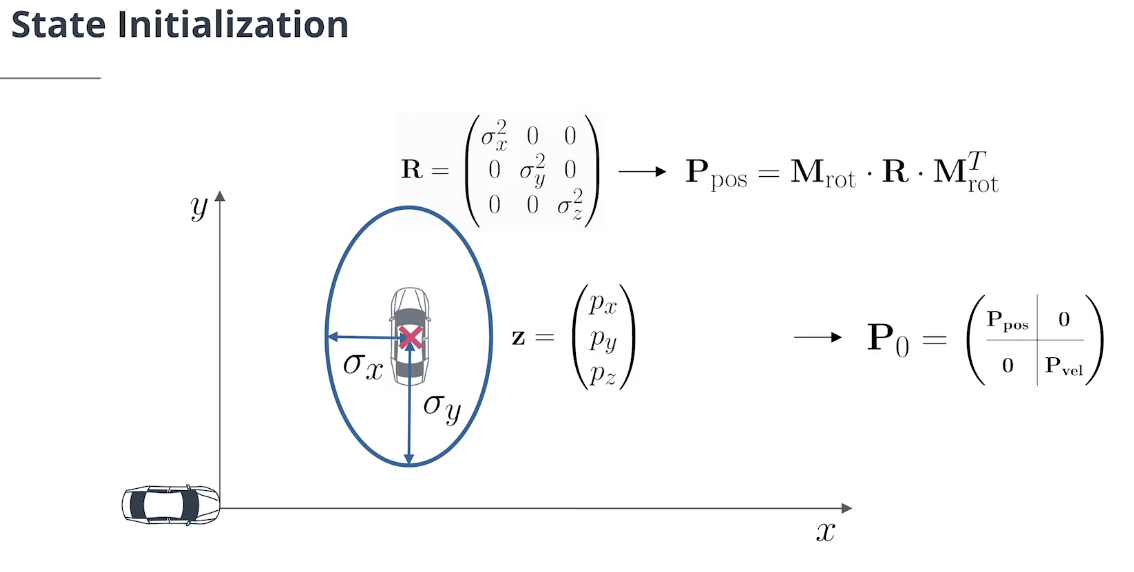
3x3的位置估计误差协方差矩阵$\mathbf p _ {\text {pos}}$可以通过从传感器坐标旋转到车辆坐标的测量协方差$\textbf R$初始化:
\[\mathbf P_{\text{pos}}= \mathbf M_{\text{rot}}\cdot \mathbf R \cdot \mathbf M_{\text{rot}}^T\]3x3的速度估计误差协方差矩阵 $\mathbf P_{\text{vel}}$ 可以使用包含较大对角值的对角矩阵初始化,因为我们无法测量速度,因此具有巨大的初始速度不确定性。
可将总体估计误差协方差初始化为:
\[\mathbf P_0 = \left(\begin{array}{c|c} \underline{\mathbf P_{\text{pos}}} & {0}\\ {0} & \overline{\mathbf P_{\text{vel}}} \end{array}\right)\]2.3 协方差旋转变换的推导
测量协方差$\text R$通过期望$E$定义:
\[\mathbf R = cov(\mathbf z) = E\left[(\mathbf z - E(\mathbf z))(\mathbf z - E(\mathbf z))^T\right]\]记住测量方程:
\[\mathbf z = h(\mathbf x)+\omega\]其中$ \omega \sim \mathcal{N}\left(0, \mathbf R\right)$是均值为0,协方差为$\text R$的测量噪声。由于ω均值为零,$\mathbf z$的期望仅为$h(\mathbf x)$:
\[E(\mathbf z) = E(h(\mathbf x)+\omega) = h(\mathbf x)\]则有:
\[\mathbf R = E\left[(\mathbf z - E(\mathbf z))(\mathbf z - E(\mathbf z))^T\right]\] \[= E\left[(\mathbf h(\mathbf x)+\omega - E(h(\mathbf x)+\omega))(...)^T\right]\] \[= E\left[(\mathbf h(\mathbf x)+\omega - h(\mathbf x))(...)^T\right]\] \[= E\left[\omega \omega^T\right]\]现在我们要将测量值从传感器旋转到车辆坐标,因此我们得到(注意$(\mathbf A\mathbf B)^T=\mathbf B^T\mathbf A^T$
\[\mathbf P_{\text{pos}} = E\left[\mathbf M_{\text{rot}}\omega \cdot(\mathbf M_{\text{rot}} \omega)^T\right]\] \[= E\left[\mathbf M_{\text{rot}}\omega \omega^T\mathbf M_{\text{rot}} ^T\right]\] \[= \mathbf M_{\text{rot}}E\left[\omega \omega^T\right]\mathbf M_{\text{rot}} ^T\] \[=\mathbf M_{\text{rot}}\cdot \mathbf R \cdot \mathbf M_{\text{rot}}^T\]类似的推理同样可以应用于卡尔曼滤波方程的其他表达式。例如:
\[\mathbf P^- = \mathbf F\mathbf P^+\mathbf F^T+\mathbf Q\] \[\mathbf S = \mathbf H\mathbf P^+\mathbf H^T+\mathbf R\]2.4 Python代码实现追踪初始化
import numpy as np
import matplotlib
matplotlib.use('wxagg') # change backend so that figure maximizing works on Mac as well
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
class Track:
'''Track class with state, covariance, id'''
def __init__(self, meas, id):
print('creating track no.', id)
self.id = id
# transform measurement to vehicle coordinates
pos_sens = np.ones((4, 1)) # homogeneous coordinates
pos_sens[0:3] = meas.z[0:3]
pos_veh = meas.sens_to_veh*pos_sens
# save initial state from measurement
self.x = np.zeros((6,1))
self.x[0:3] = pos_veh[0:3]
# set up position estimation error covariance
M_rot = meas.sens_to_veh[0:3, 0:3]
P_pos = M_rot * meas.R * np.transpose(M_rot)
# set up velocity estimation error covariance
sigma_p44 = 50 # initial setting for estimation error covariance P entry for vx
sigma_p55 = 50 # initial setting for estimation error covariance P entry for vy
sigma_p66 = 5 # initial setting for estimation error covariance P entry for vz
P_vel = np.matrix([[sigma_p44**2, 0, 0],
[0, sigma_p55**2, 0],
[0, 0, sigma_p66**2]])
# overall covariance initialization
self.P = np.zeros((6, 6))
self.P[0:3, 0:3] = P_pos
self.P[3:6, 3:6] = P_vel
###################
class Measurement:
'''Lidar measurement class including measurement z, covariance R, coordinate transform matrix'''
def __init__(self, gt, phi, t):
# compute rotation around z axis
M_rot = np.matrix([[np.cos(phi), -np.sin(phi), 0],
[np.sin(phi), np.cos(phi), 0],
[0, 0, 1]])
# coordiante transformation matrix from sensor to vehicle coordinates
self.sens_to_veh = np.matrix(np.identity(4))
self.sens_to_veh[0:3, 0:3] = M_rot
self.sens_to_veh[0:3, 3] = t
print('Coordinate transformation matrix:', self.sens_to_veh)
# transform ground truth from vehicle to sensor coordinates
gt_veh = np.ones((4, 1)) # homogeneous coordinates
gt_veh[0:3] = gt[0:3]
gt_sens = np.linalg.inv(self.sens_to_veh) * gt_veh
# create measurement object
sigma_lidar_x = 0.01 # standard deviation for noisy measurement generation
sigma_lidar_y = 0.01
sigma_lidar_z = 0.001
self.z = np.zeros((3,1)) # measurement vector
self.z[0] = float(gt_sens[0,0]) + np.random.normal(0, sigma_lidar_x)
self.z[1] = float(gt_sens[1,0]) + np.random.normal(0, sigma_lidar_y)
self.z[2] = float(gt_sens[2,0]) + np.random.normal(0, sigma_lidar_z)
self.R = np.matrix([[sigma_lidar_x**2, 0, 0], # measurement noise covariance matrix
[0, sigma_lidar_y**2, 0],
[0, 0, sigma_lidar_z**2]])
def visualize(track, meas):
fig, (ax1, ax2, ax3) = plt.subplots(1,3)
ax1.scatter(-meas.z[1], meas.z[0], marker='o', color='blue', label='measurement')
ax2.scatter(-track.x[1], track.x[0], color='red', s=80, marker='x', label='initialized track')
# transform measurement to vehicle coordinates for visualization
z_sens = np.ones((4, 1)) # homogeneous coordinates
z_sens[0:3] = meas.z[0:3]
z_veh = meas.sens_to_veh * z_sens
ax3.scatter(-float(z_veh[1]), float(z_veh[0]), marker='o', color='blue', label='measurement')
ax3.scatter(-track.x[1], track.x[0], color='red', s=80, marker='x', label='initialized track')
# maximize window
mng = plt.get_current_fig_manager()
mng.frame.Maximize(True)
# legend and axes
for ax in (ax1, ax2, ax3):
ax.legend(loc='center left', shadow=True, fontsize='large', bbox_to_anchor=(0.5, 0.1))
ax.set_xlabel('y [m]')
ax.set_ylabel('x [m]')
ax.set_xlim(-2,2)
ax.set_ylim(0,2)
# correct x ticks (positive to the left)
ticks_x = ticker.FuncFormatter(lambda x, pos: '{0:g}'.format(-x) if x!=0 else '{0:g}'.format(x))
ax.xaxis.set_major_formatter(ticks_x)
# titles
ax1.title.set_text('Sensor Coordinates')
ax2.title.set_text('Vehicle Coordinates')
ax3.title.set_text('Vehicle Coordinates\n (track and measurement should align)')
plt.show()
###################
# ground truth
gt = np.matrix([[1.7],
[1],
[0]])
# sensor translation and rotation angle
t = np.matrix([[2],
[0.5],
[0]])
phi = np.radians(45)
# generate measurement
meas = Measurement(gt, phi, t)
# initialize track from this measurement
track = Track(meas, 1)
# visualization
visualize(track, meas)
3. 追踪状态管理
3.1 追踪分数评估
我们可以通过设置来定义一个简单的追踪分数:
\[\text{score} = \frac{\#\text{detections in last $n$ frames}}{n}\]有许多其他的可能性来定义一个跟踪分数,信心或存在的概率。
基于追踪分数,可以定义追踪状态,例如“已初始化”、“暂定”或“已确认”。
示例跟踪删除条件
下面是一个启发式轨迹删除标准的示例:
- 确认追踪:
如果分数<0.6,则删除。这只适用于已确认的追踪,因此之前的分数必须高于0.8,然后降至0.6以下。
如果$\mathbf P_{11}<3^2$或$\mathbf P_{22}<3^2$则删除。这意味着我们的目标可能在半径为3米圆内的任何地方,位置不确定性增加了很多,因此追踪应该被删除。
- 暂定或初始化追踪:
如果分数<0.17,则删除。请注意,此阈值远低于已确认追踪的阈值,我们不希望在新追踪稳定之前立即删除它们。
如果$\mathbf P_{11}<3^2$,或$\mathbf P_{22}<3^2$则删除。
3.2 可见度
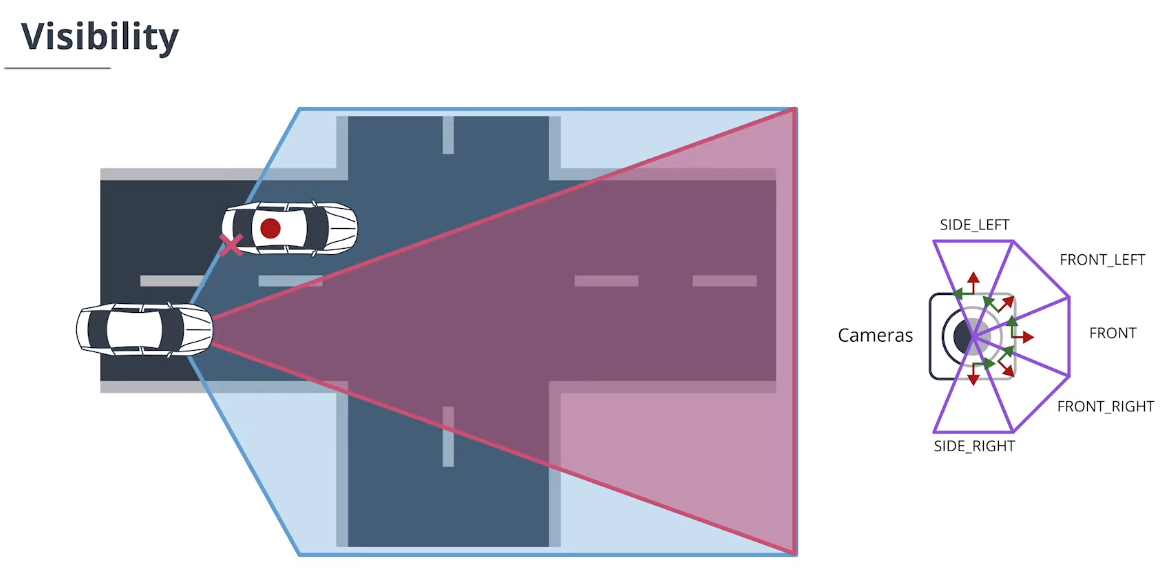
在传感器融合系统中,每个传感器通常具有不同的视野。我们需要一个检测概率或可见性推理来避免追踪分数振荡。
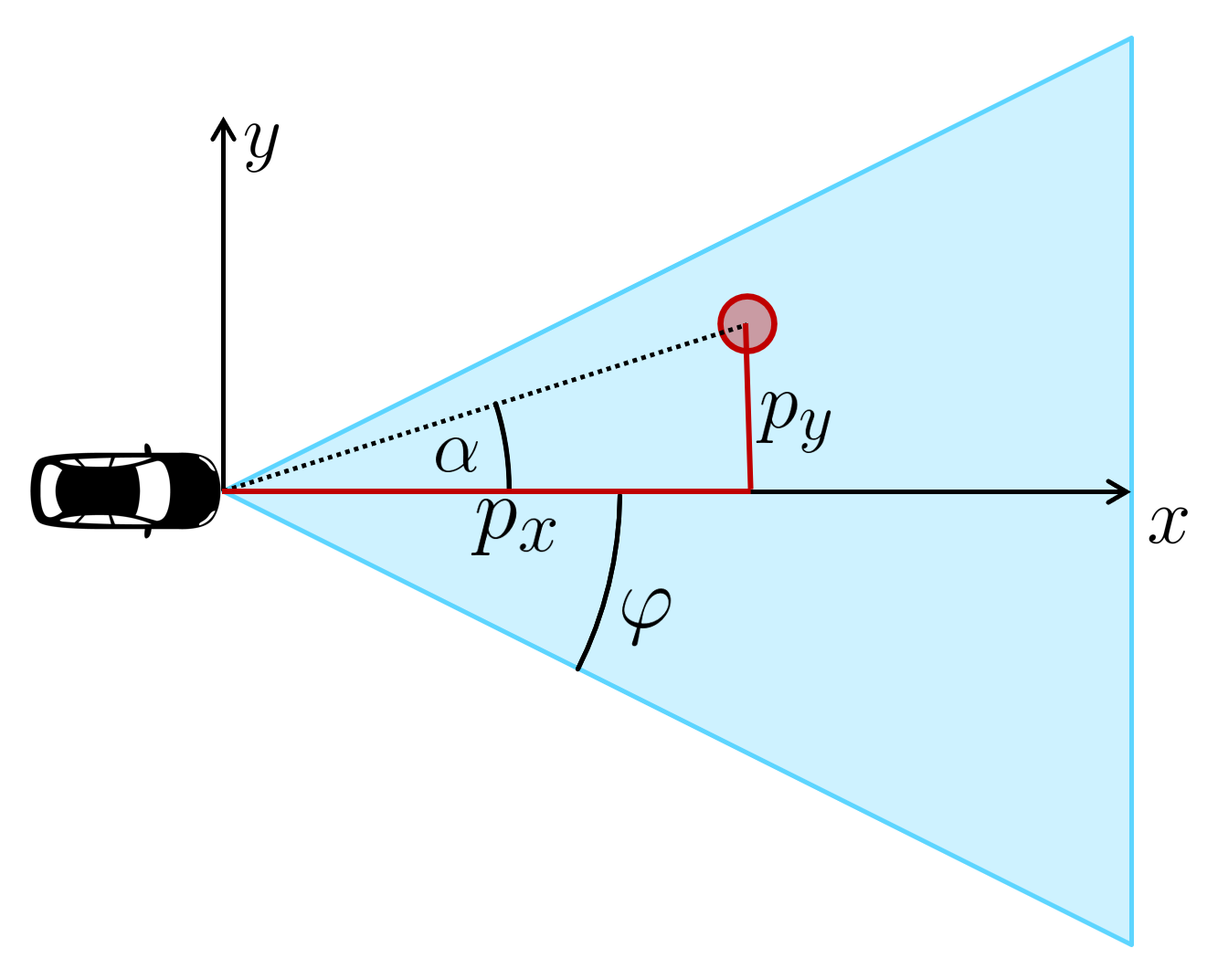
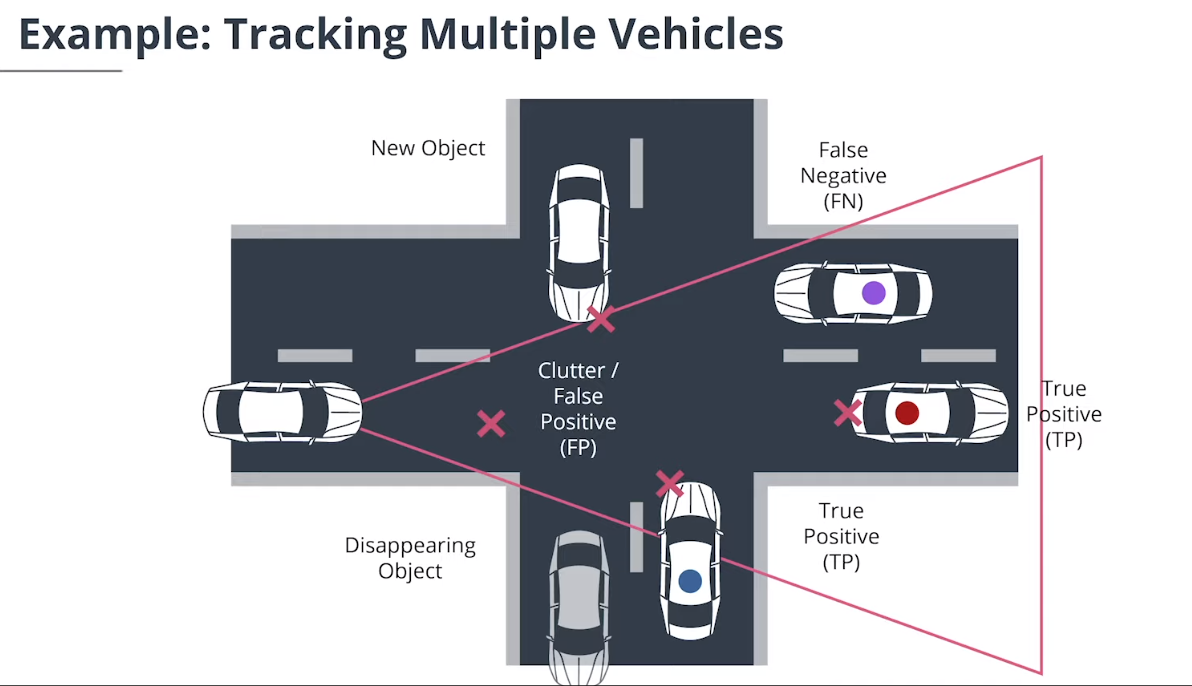
看看这张图片。我们想检查红色物体的可见性,它的坐标$p_x,p_y$已转换为此图像中的传感器坐标。我们必须计算红色目标相对于车辆的角度$α$。如果α的绝对值小于相机的开放角度$φ$,我们可以看到物体,否则我们看不到。那么我们如何计算α?
可以用三角函数计算$α$。我们已经知道对边$p_y$和邻边$p_x$,我们假设$p_x$>0,因此可以使用正切函数:
\[\tan(\alpha) = \frac{p_y}{p_x}\]通过使用反正切函数即可求得所需的角度$\alpha$:
\[\alpha = \arctan\left(\frac{p_y}{p_x}\right)\]3.3 Python实现相机追踪可视角度推理
# imports
import numpy as np
import matplotlib
#matplotlib.use('wxagg') # change backend so that figure maximizing works on Mac as well
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
class Camera:
'''Camera sensor class including field of view and coordinate transformation'''
def __init__(self, phi, t):
self.fov = [-np.pi/4, np.pi/4] # sensor field of view / opening angle
# compute rotation around z axis
M_rot = np.matrix([[np.cos(phi), -np.sin(phi), 0],
[np.sin(phi), np.cos(phi), 0],
[0, 0, 1]])
print(M_rot)
# coordiante transformation matrix from sensor to vehicle coordinates
self.sens_to_veh = np.matrix(np.identity(4))
self.sens_to_veh[0:3, 0:3] = M_rot
self.sens_to_veh[0:3, 3] = t
self.veh_to_sens = np.linalg.inv(self.sens_to_veh) # transformation vehicle to sensor coordinates
def in_fov(self, x):
# check if an object x can be seen by this sensor
pos_veh = np.ones((4, 1)) # homogeneous coordinates
pos_veh[0:3] = x[0:3]
pos_sens = self.veh_to_sens*pos_veh # transform from vehicle to sensor coordinates
visible = False
# make sure to not divide by zero - we can exclude the whole negative x-range here
if pos_sens[0] > 0:
alpha = np.arctan(pos_sens[1]/pos_sens[0]) # calc angle between object and x-axis
# no normalization needed because returned alpha always lies between [-pi/2, pi/2]
if alpha > self.fov[0] and alpha < self.fov[1]:
visible = True
return visible
#################
def run():
'''generate random points and check visibility'''
# camera with translation and rotation angle
t = np.matrix([[2],
[0],
[0]])
phi = np.radians(45)
cam = Camera(phi, t)
# initialize visualization
fig, ax = plt.subplots()
for i in range(50):
# define track position and velocity
x = np.matrix([[np.random.uniform(-5,5)],
[np.random.uniform(-5,5)],
[0],
[0],
[0],
[0]])
# check if x is visible by camera
result = cam.in_fov(x)
# plot results
pos_veh = np.ones((4, 1)) # homogeneous coordinates
pos_veh[0:3] = x[0:3]
pos_sens = cam.veh_to_sens*pos_veh # transform from vehicle to sensor coordinates
if result == True:
col = 'green'
ax.scatter(float(-pos_sens[1]), float(pos_sens[0]), marker='o', color=col, label='visible track')
else:
col = 'red'
ax.scatter(float(-pos_sens[1]), float(pos_sens[0]), marker='o', color=col, label='invisible track')
ax.text(float(-pos_sens[1]), float(pos_sens[0]), str(result))
# plot FOV
ax.plot([0, -5], [0, 5], color='blue', label='field of view')
ax.plot([0, 5], [0, 5], color='blue')
# maximize window
mng = plt.get_current_fig_manager()
#mng.frame.Maximize(True)
# remove repeated labels
handles, labels = ax.get_legend_handles_labels()
handle_list, label_list = [], []
for handle, label in zip(handles, labels):
if label not in label_list:
handle_list.append(handle)
label_list.append(label)
ax.legend(handle_list, label_list, loc='center left', shadow=True, fontsize='large', bbox_to_anchor=(0.9, 0.1))
# axis
ax.set_xlabel('y [m]')
ax.set_ylabel('x [m]')
ax.set_xlim(-5,5)
ax.set_ylim(0,5)
# correct x ticks (positive to the left)
ticks_x = ticker.FuncFormatter(lambda x, pos: '{0:g}'.format(-x) if x!=0 else '{0:g}'.format(x))
ax.xaxis.set_major_formatter(ticks_x)
plt.show()
####################
# call main loop
run()
4. 数据关联
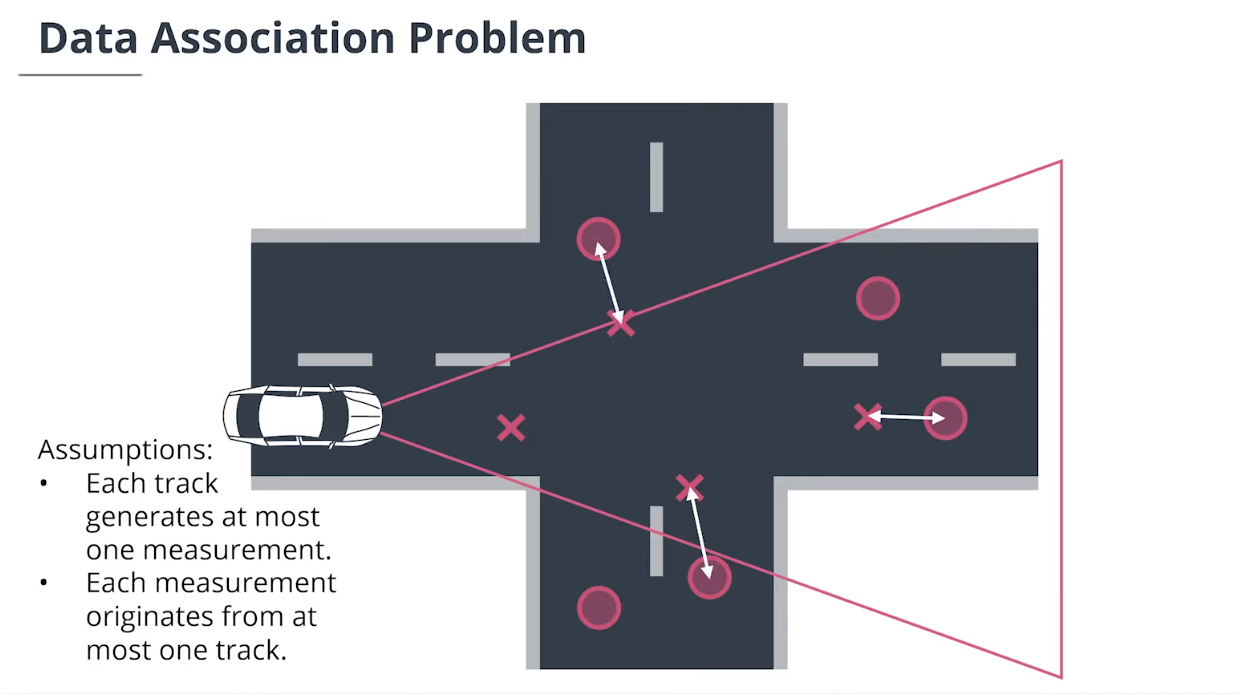
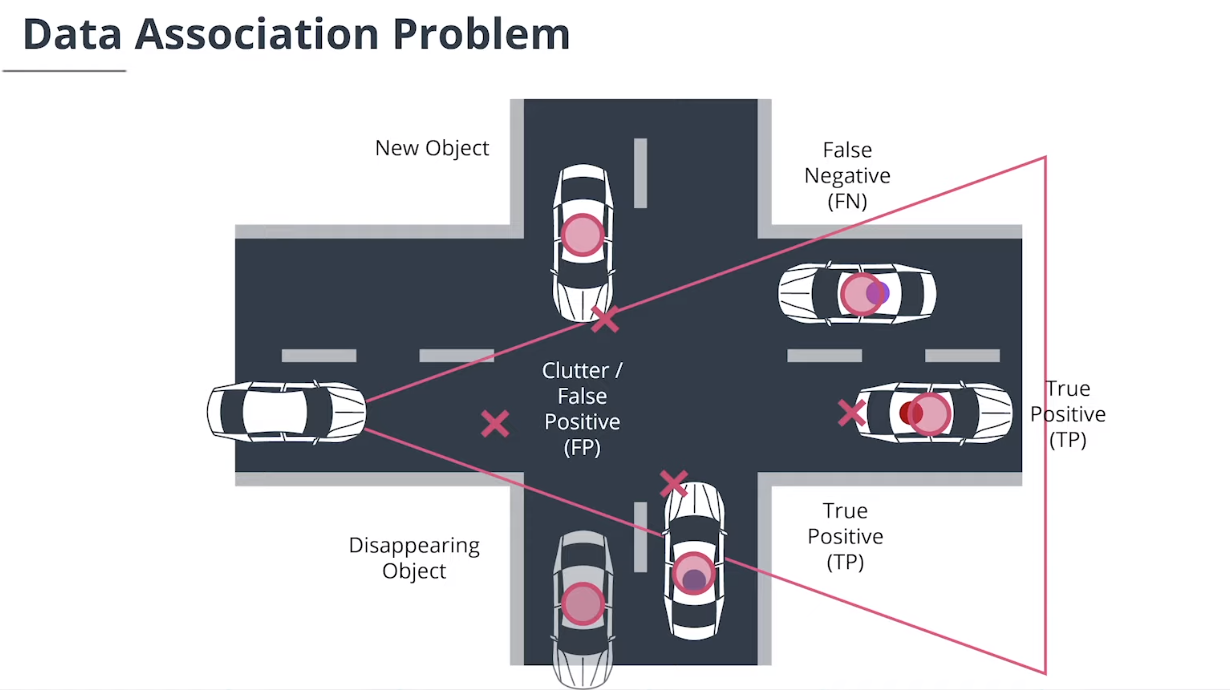
$\textbf{data association}$数据关联模块分配测量值给追踪,并决定哪个跟踪使用哪个测量值。
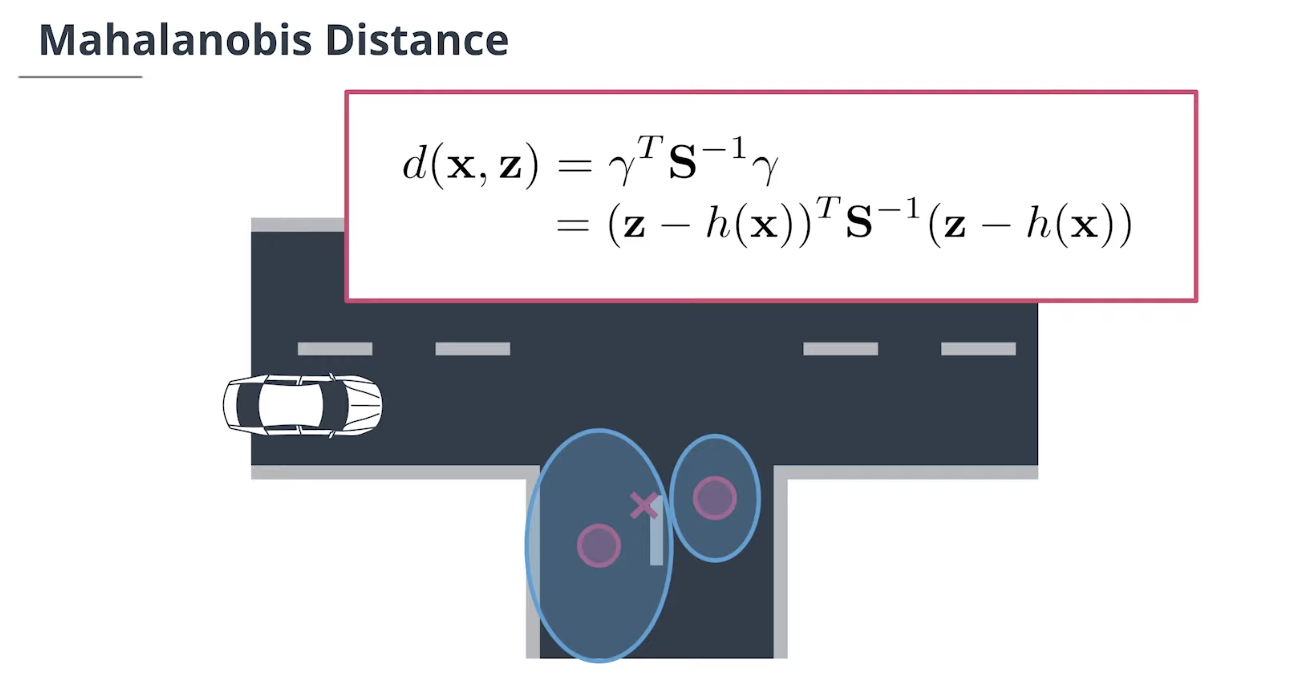
$\textbf{data association}$数据关联模块主要使用$\textbf{Mahalanobis distance}$马氏距离来做决定。
\[d(\mathbf x, \mathbf z)= \gamma^T\mathbf S^{-1}\gamma = (\mathbf z - h(\mathbf x))^T\mathbf S^{-1}(\mathbf z - h(\mathbf x))\]4.1 数据关联方法概述
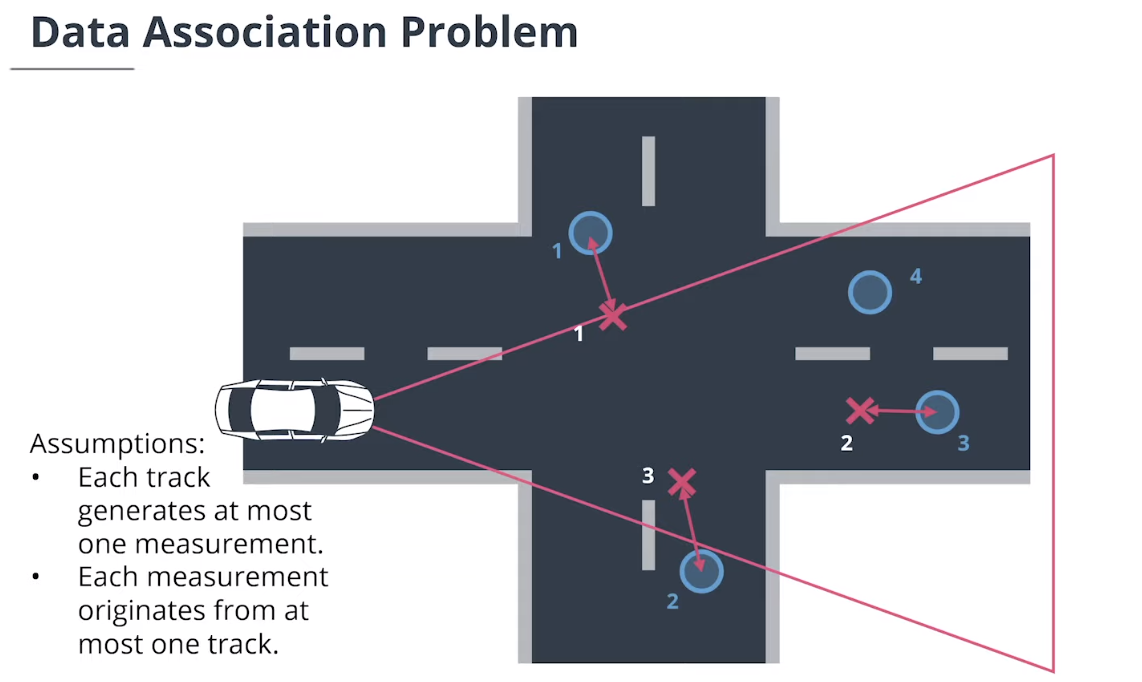
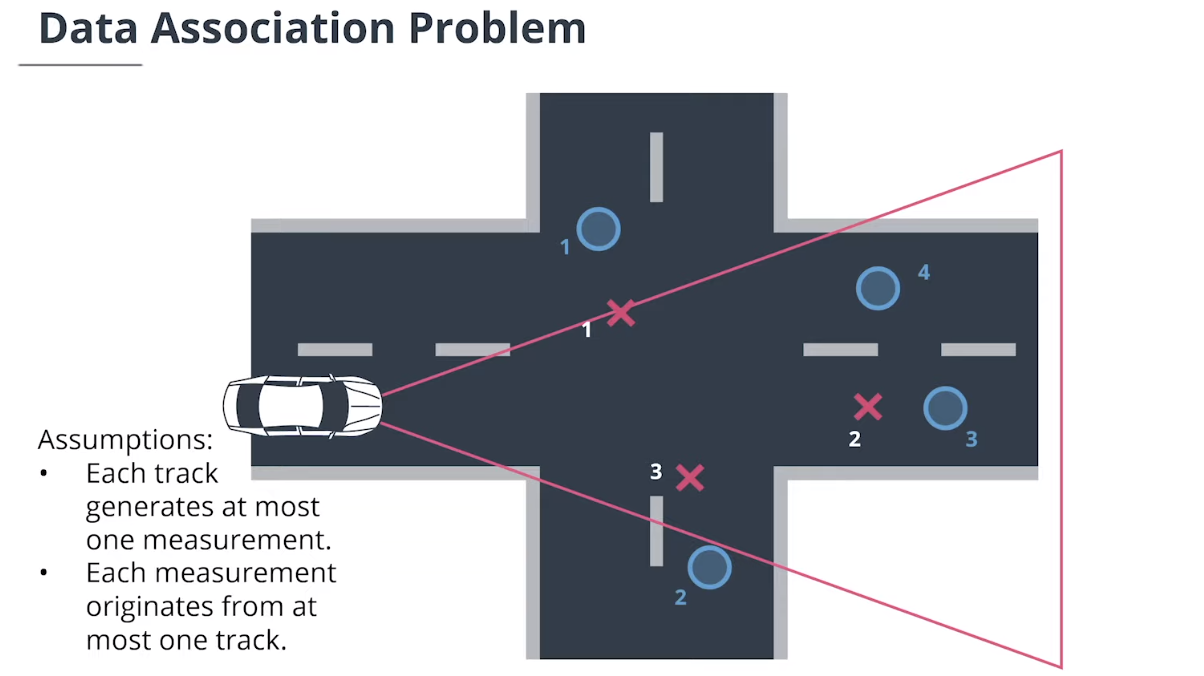
$\textbf{Simple Nearest Neighbor(SNN)}$简单最近邻数据关联计算追踪和测量值之间的所有马氏距离,然后迭代更新最近关联对。
SNN不是全局最优的,这可以使用$\textbf{Global Nearest Neighbor(GNN)}$全局最近邻数据关联来解决。
这两种数据关联方法都在不明确的情况下强制执行硬决策。更高级的$\textbf{Probabilistic Data Association(PDA)}$概率数据关联(PDA)技术可以避免这些困难的决策和由此产生的错误。此类有不同的变体(例如PDA、JPDA、JIPDA)。
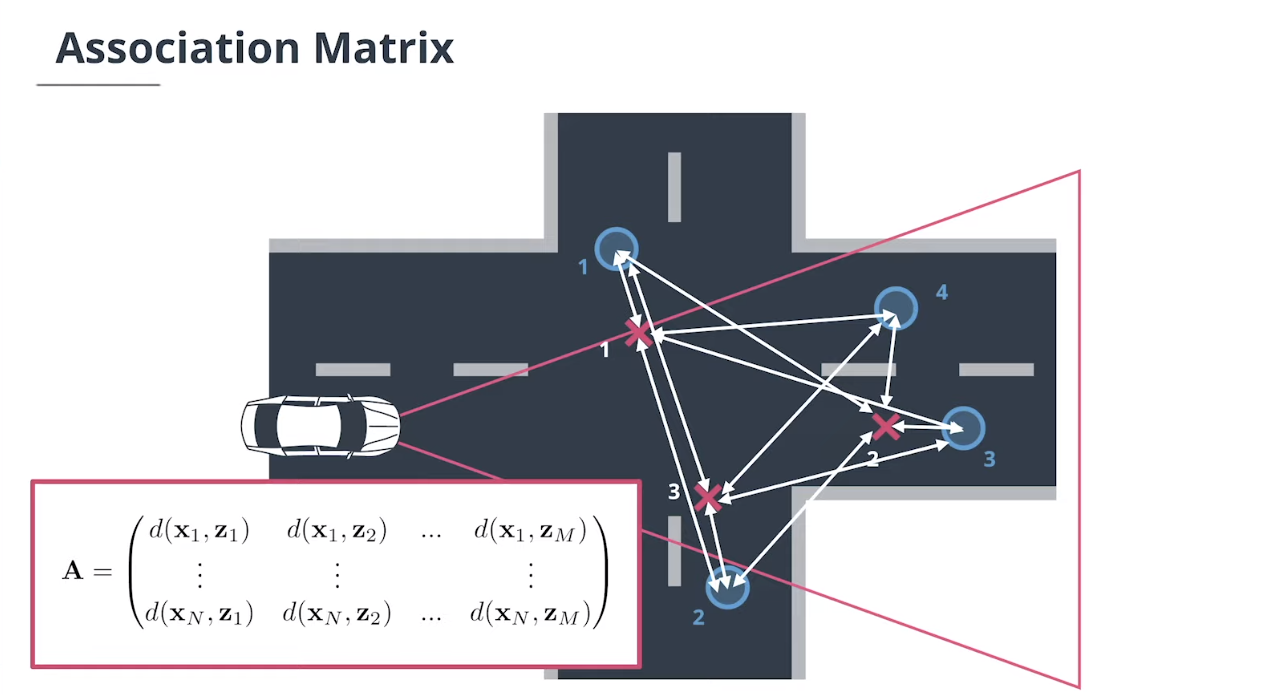
4.2 数据关联矩阵
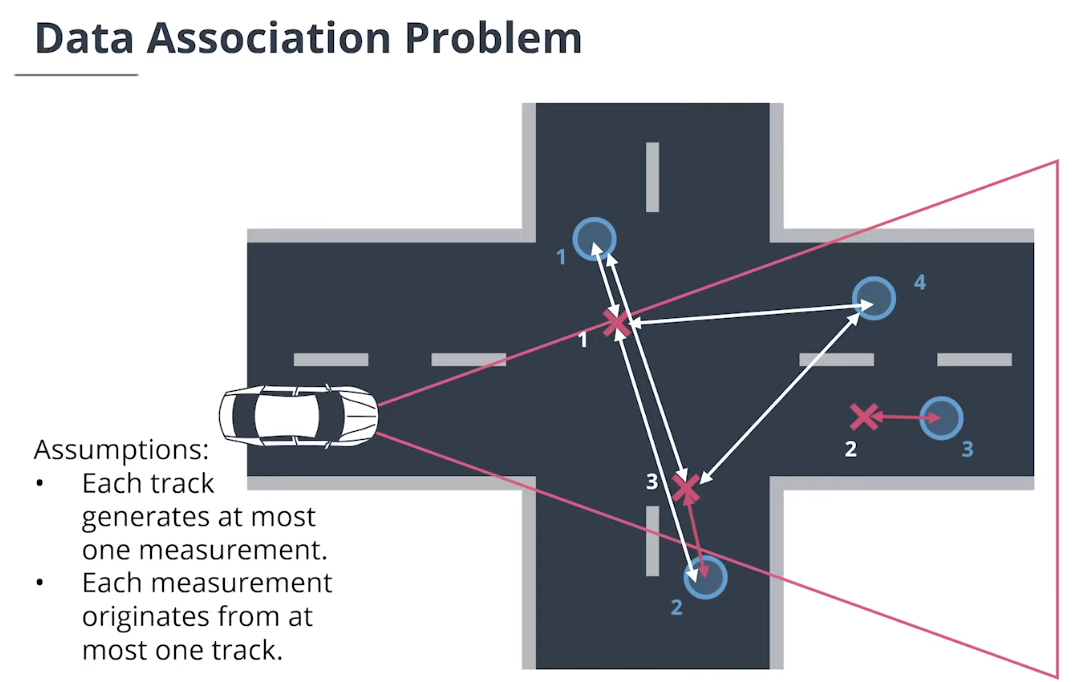
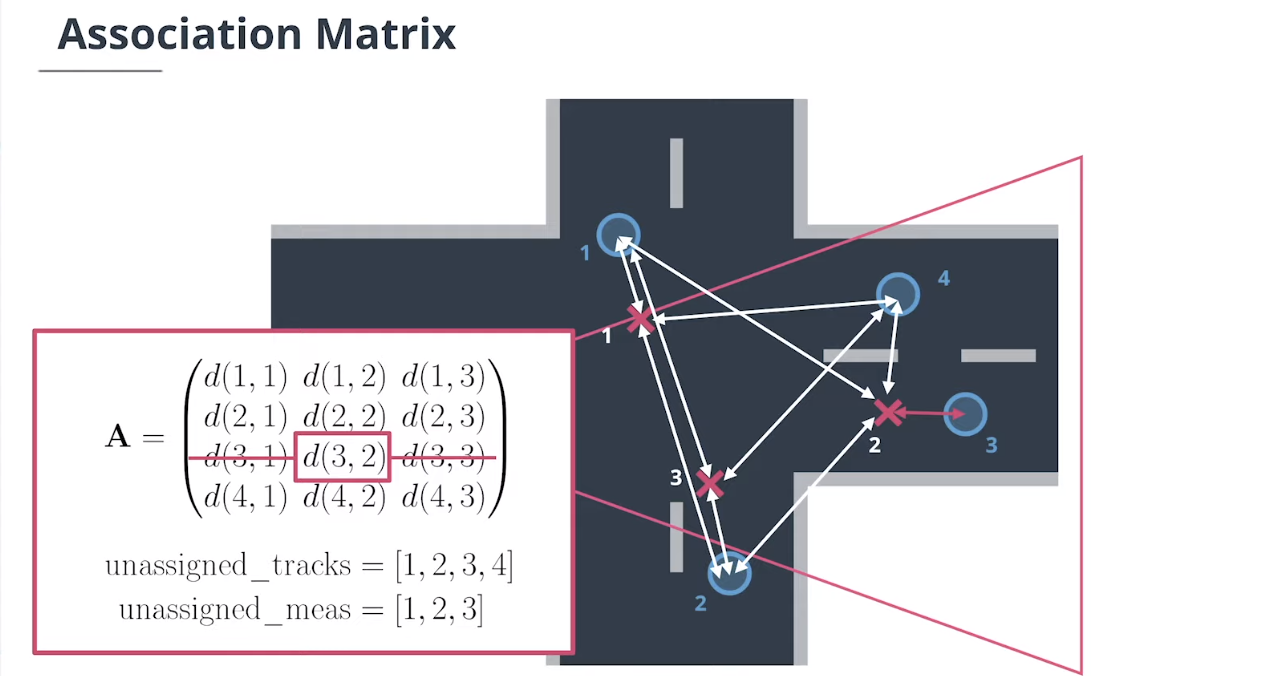
假设我们有$N$个追踪目标轨迹和$M$个测量值。
$\textbf{association matrix}$关联矩阵$\mathbf A$是一个$N×M$的矩阵,它包含每个追踪和每个测量之间的马氏距离:
\[\mathbf A = \begin{pmatrix} d(\mathbf x_1, \mathbf z_1) & d(\mathbf x_1, \mathbf z_2) &...&d(\mathbf x_1, \mathbf z_M)\\ \vdots &\vdots&&\vdots\\ d(\mathbf x_N, \mathbf z_1) & d(\mathbf x_N, \mathbf z_2) &...&d(\mathbf x_N, \mathbf z_M) \end{pmatrix}\]我们还需要一个未指定追踪的列表和一个未指定测量的列表。
我们在$A$中查找最小的条目,以确定使用哪个度量更新哪个追踪,然后从$A$中删除该行和列,并从列表中删除追踪ID和度量ID。我们重复这个过程,直到$A$为空。
4.3 Python实现数据关联矩阵
import numpy as np
import matplotlib
matplotlib.use('wxagg') # change backend so that figure maximizing works on Mac as well
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
class Association:
'''Data association class with single nearest neighbor association and gating based on Mahalanobis distance'''
def __init__(self):
self.association_matrix = np.matrix([])
def associate(self, track_list, meas_list):
N = len(track_list) # N tracks
M = len(meas_list) # M measurements
# initialize association matrix
self.association_matrix = np.inf*np.ones((N,M))
# loop over all tracks and all measurements to set up association matrix
for i in range(N):
track = track_list[i]
for j in range(M):
meas = meas_list[j]
dist = self.MHD(track, meas)
self.association_matrix[i,j] = dist
def MHD(self, track, meas):
# calc Mahalanobis distance
H = np.matrix([[1, 0, 0, 0],
[0, 1, 0, 0]])
gamma = meas.z - H*track.x
S = H*track.P*H.transpose() + meas.R
MHD = gamma.transpose()*np.linalg.inv(S)*gamma # Mahalanobis distance formula
return MHD
##################
class Track:
'''Track class with state, covariance, id'''
def __init__(self, id):
# save id
self.id = id
# generate random state x
self.x = np.matrix([[np.random.uniform(2,8)],
[np.random.uniform(-3,3)],
[0],
[0]])
# set up estimation error covariance
self.P = np.matrix([[2, 0, 0, 0],
[0, 3, 0, 0],
[0, 0, 1, 0],
[0, 0, 0, 1]])
class Measurement:
'''Measurement class with easurement, covariance, id'''
def __init__(self, id, x, y):
# save id
self.id = id
# generate random measurement z
self.z = np.matrix([[x + np.random.normal(0,2)],
[y + np.random.normal(0,2)]])
# set up measurement covariance
self.R = np.matrix([[2, 0],
[0, 2]])
#################
def run():
'''generate tracks and measurements and call association'''
# set up track_list and meas_list for association
np.random.seed(5) # make random values predictable
association = Association() # init data association
track_list = []
meas_list = []
# initialize visualization
fig, ax = plt.subplots()
# generate and plot tracks and measurements
for i in range(3):
# tracks
track = Track(i+1)
track_list.append(track)
ax.scatter(float(-track.x[1]), float(track.x[0]), marker='x', color='red', label='track')
ax.text(float(-track.x[1]), float(track.x[0]), str(track.id), color='red')
# measurements
meas = Measurement(i+1, float(track.x[0]), float(track.x[1]))
meas_list.append(meas)
ax.scatter(float(-meas.z[1]), float(meas.z[0]), marker='o', color='green', label='measurement')
ax.text(float(-meas.z[1]), float(meas.z[0]), str(meas.id), color='green')
# calculate association matrix
association.associate(track_list, meas_list)
print('Association matrix:', association.association_matrix)
# visualize distances
for track in track_list:
for meas in meas_list:
dist = association.association_matrix[track.id-1, meas.id-1]
if dist < np.inf:
ax.plot([float(-track.x[1]), float(-meas.z[1])], [float(track.x[0]), float(meas.z[0])], color='gray')
str_dist = "{:.2f}".format(dist)
ax.text(float((-track.x[1] - meas.z[1])/2), float((track.x[0] + meas.z[0])/2), str_dist)
# maximize window
mng = plt.get_current_fig_manager()
mng.frame.Maximize(True)
# remove repeated labels
handles, labels = ax.get_legend_handles_labels()
handle_list, label_list = [], []
for handle, label in zip(handles, labels):
if label not in label_list:
handle_list.append(handle)
label_list.append(label)
ax.legend(handle_list, label_list, loc='center left', shadow=True, fontsize='large', bbox_to_anchor=(0.9, 0.1))
# axis
ax.set_xlabel('y [m]')
ax.set_ylabel('x [m]')
ax.set_xlim(-5,5)
ax.set_ylim(0,10)
# correct x ticks (positive to the left)
ticks_x = ticker.FuncFormatter(lambda x, pos: '{0:g}'.format(-x) if x!=0 else '{0:g}'.format(x))
ax.xaxis.set_major_formatter(ticks_x)
plt.show()
####################
# call main loop
run()
4.4 门控技术(Gating)
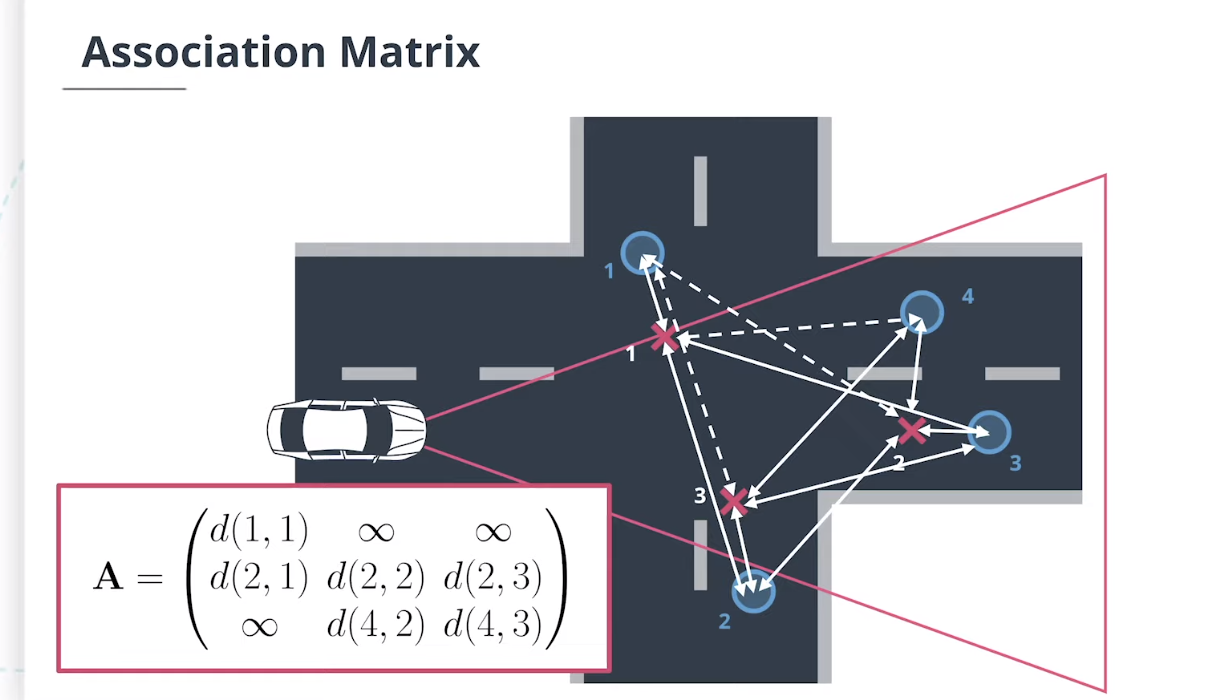
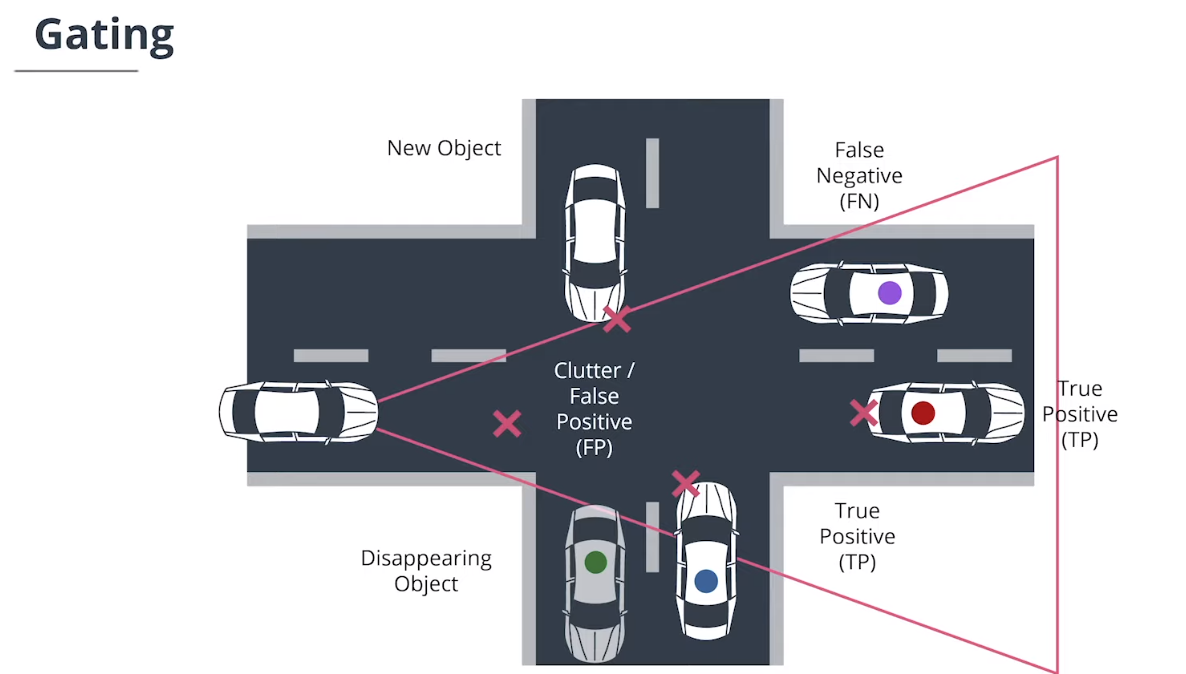
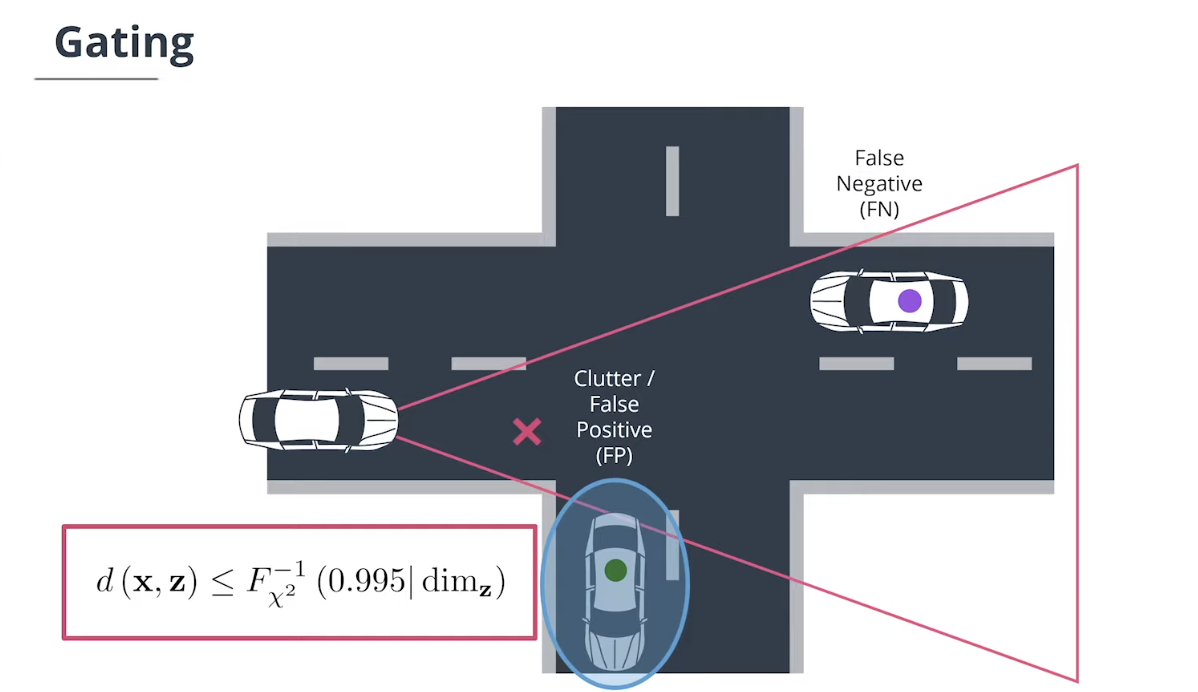
$Gating$门控技术通过删除不太可能的关联对来降低关联复杂性。
因为残差是高斯分布的,所以马氏距离遵循$\chi^2$分布。如果马氏距离小于通过逆累积$\chi^2$分布计算得出的阈值,则测量值位于追踪门内,阈值计算方式如下:
\[d\left(\mathbf x,\mathbf z\right)\leq F_{\chi^2}^{-1}\left(0.995|\dim_{\mathbf z}\right)\]如果测量值位于追踪门控外,我们可以将其马氏距离设置为无穷大,例如:
\[\mathbf A = \begin{pmatrix} d(1, 1) & \infty &\infty \\ d(2, 1) & d(2, 2) &d(2, 3) \\ d(3, 1) & d(3, 2) &d(3, 3) \\ \infty& d(4, 2) &d(4, 3) \end{pmatrix}\]| 计算逆累积$\chi^2$分布 $F_{\chi^2}^{-1}\left(p | df\right)$使用概率$p$和自由度$df$(等于此处测量空间的尺寸),可以使用scipy软件包: |
from scipy.stats.distributions import chi2
chi2.ppf(p, df)
ppf代表百分点函数,它是累积密度函数的倒数。有关更多详细信息,请参阅相关的scipy文档。
要获取关联矩阵$A$中最小项的索引,可以使用numpy.argmin。通常,此函数返回展平数组的索引。为了避免这种情况,我们可以使用numpy.unravel_index,如下所示:
np.unravel_index(np.argmin(A, axis=None), A.shape)
4.5 Python实现数据关联门控技术
import numpy as np
import matplotlib
matplotlib.use('wxagg') # change backend so that figure maximizing works on Mac as well
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
from scipy.stats.distributions import chi2
class Association:
'''Data association class with single nearest neighbor association and gating based on Mahalanobis distance'''
def __init__(self):
self.association_matrix = np.matrix([])
self.unassigned_tracks = []
self.unassigned_meas = []
def associate(self, track_list, meas_list):
N = len(track_list) # N tracks
M = len(meas_list) # M measurements
self.unassigned_tracks = list(range(N))
self.unassigned_meas = list(range(M))
# initialize association matrix
self.association_matrix = np.inf*np.ones((N,M))
# loop over all tracks and all measurements to set up association matrix
for i in range(N):
track = track_list[i]
for j in range(M):
meas = meas_list[j]
dist = self.MHD(track, meas)
if self.gating(dist):
self.association_matrix[i,j] = dist
def MHD(self, track, meas):
# calc Mahalanobis distance
H = np.matrix([[1, 0, 0, 0],
[0, 1, 0, 0]])
gamma = meas.z - H*track.x
S = H*track.P*H.transpose() + meas.R
MHD = gamma.transpose()*np.linalg.inv(S)*gamma # Mahalanobis distance formula
return MHD
def gating(self, MHD):
# check if measurement lies inside gate
limit = chi2.ppf(0.95, df=2)
if MHD < limit:
return True
else:
return False
def get_closest_track_and_meas(self):
# find closest track and measurement for next update
A = self.association_matrix
if np.min(A) == np.inf:
return np.nan, np.nan
# get indices of minimum entry
ij_min = np.unravel_index(np.argmin(A, axis=None), A.shape)
ind_track = ij_min[0]
ind_meas = ij_min[1]
# delete row and column for next update
A = np.delete(A, ind_track, 0)
A = np.delete(A, ind_meas, 1)
self.association_matrix = A
# update this track with this measurement
update_track = self.unassigned_tracks[ind_track]
update_meas = self.unassigned_meas[ind_meas]
# remove this track and measurement from list
self.unassigned_tracks.remove(update_track)
self.unassigned_meas.remove(update_meas)
return update_track, update_meas
##################
class Track:
'''Track class with state, covariance, id'''
def __init__(self, id):
# save id
self.id = id
# generate random state x
self.x = np.matrix([[np.random.uniform(2,8)],
[np.random.uniform(-3,3)],
[0],
[0]])
# set up estimation error covariance
self.P = np.matrix([[2, 0, 0, 0],
[0, 3, 0, 0],
[0, 0, 1, 0],
[0, 0, 0, 1]])
class Measurement:
'''Measurement class with easurement, covariance, id'''
def __init__(self, id, x, y):
# save id
self.id = id
# generate random measurement z
self.z = np.matrix([[x + np.random.normal(0,2)],
[y + np.random.normal(0,2)]])
# set up measurement covariance
self.R = np.matrix([[2, 0],
[0, 2]])
#################
def run():
'''generate tracks and measurements and call association'''
# set up track_list and meas_list for association
np.random.seed(5) # make random values predictable
association = Association() # init data association
track_list = []
meas_list = []
# initialize visualization
fig, ax = plt.subplots()
# generate and plot tracks and measurements
for i in range(3):
# tracks
track = Track(i+1)
track_list.append(track)
ax.scatter(float(-track.x[1]), float(track.x[0]), marker='x', color='red', label='track')
ax.text(float(-track.x[1]), float(track.x[0]), str(track.id), color='red')
# measurements
meas = Measurement(i+1, float(track.x[0]), float(track.x[1]))
meas_list.append(meas)
ax.scatter(float(-meas.z[1]), float(meas.z[0]), marker='o', color='green', label='measurement')
ax.text(float(-meas.z[1]), float(meas.z[0]), str(meas.id), color='green')
# calculate association matrix
association.associate(track_list, meas_list)
print('Association matrix:', association.association_matrix)
print('unassigned_tracks list:', association.unassigned_tracks)
print('unassigned_meas list:', association.unassigned_meas)
# visualize distances
for track in track_list:
for meas in meas_list:
dist = association.association_matrix[track.id-1, meas.id-1]
if dist < np.inf:
ax.plot([float(-track.x[1]), float(-meas.z[1])], [float(track.x[0]), float(meas.z[0])], color='gray')
str_dist = "{:.2f}".format(dist)
ax.text(float((-track.x[1] - meas.z[1])/2), float((track.x[0] + meas.z[0])/2), str_dist)
# update associated tracks with measurements
matrix_orig = association.association_matrix
while association.association_matrix.shape[0]>0 and association.association_matrix.shape[1]>0:
# search for next association between a track and a measurement
ind_track, ind_meas = association.get_closest_track_and_meas()
if np.isnan(ind_track):
print('---no more associations---')
break
track = track_list[ind_track]
meas = meas_list[ind_meas]
dist = matrix_orig[ind_track, ind_meas]
ax.plot([float(-track.x[1]), float(-meas.z[1])], [float(track.x[0]), float(meas.z[0])], color='blue', label='association')
str_dist = "{:.2f}".format(dist)
ax.text(float((-track.x[1] - meas.z[1])/2), float((track.x[0] + meas.z[0])/2), str_dist)
print('found association between track', ind_track+1, 'and measurement', ind_meas+1, 'with MHD =', str_dist)
print('New association matrix:', association.association_matrix)
print('New unassigned_tracks list:', association.unassigned_tracks)
print('New unassigned_meas list:', association.unassigned_meas)
#################
# visualization
# maximize window
mng = plt.get_current_fig_manager()
mng.frame.Maximize(True)
# remove repeated labels
handles, labels = ax.get_legend_handles_labels()
handle_list, label_list = [], []
for handle, label in zip(handles, labels):
if label not in label_list:
handle_list.append(handle)
label_list.append(label)
ax.legend(handle_list, label_list, loc='center left', shadow=True, fontsize='large', bbox_to_anchor=(0.9, 0.1))
# axis
ax.set_xlabel('y [m]')
ax.set_ylabel('x [m]')
ax.set_xlim(-5,5)
ax.set_ylim(0,10)
# correct x ticks (positive to the left)
ticks_x = ticker.FuncFormatter(lambda x, pos: '{0:g}'.format(-x) if x!=0 else '{0:g}'.format(x))
ax.xaxis.set_major_formatter(ticks_x)
plt.show()
####################
# call main loop
run()
5. 评估跟踪性能
为了评估跟踪性能,我们可以使用$\textbf{Root Mean Square Error(RMSE)}$均方根误差,它计算估计状态和基本真值状态之间的残差:
\[RMSE = \sqrt{\frac 1n\sum_{k=1}^n \left(x_k^{\text{est}}-x_k^{\text{true}}\right)^2}\]